Human element [ hs2579 ]
Position: chr20:21,806,104-21,809,344 (UCSC browser)
Source: Lawrence Berkeley National Laboratory
Flanking genes:
PAX1
-
LOC100270679
Timepoints:
e11.5
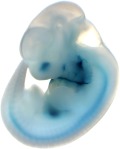
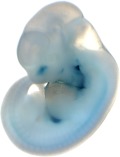
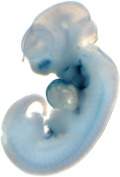
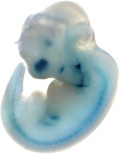
Reproducible staining in unidentified abdominal structure.
Expression Pattern
| branchial arch (8 out of 8 embryos) | | facial mesenchyme (8 out of 8 embryos) | | nose (8 out of 8 embryos) | | other (8 out of 8 embryos) |
Embryo 1
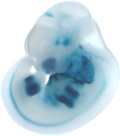
| Embryo 2
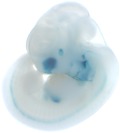
| Embryo 4

| Embryo 8
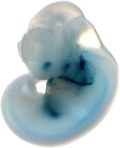
| Embryo 9
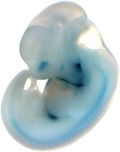
| Embryo 10
| Embryo 11
| Embryo 12
| Embryo 13
| Embryo 14
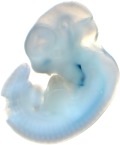
| Embryo 15
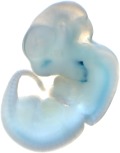
|
Fasta sequence
AAGCGACTTTTGTGCCTCAGcctcctgagtagctgggactataggtgcaagttaccacgc
cctgctaatatttttgtatttttaatagagacagggtttcaccatgttggccaggctggt
ctcgaattcctgacctcaagtaatccacccacctcagcctccaaaagtcctgggattaca
ggcgtgacccactgtgcccagctcccaaaatgtataactaagtgagaagggaaggatgta
gaatagggtgggtgttttgctattttcttgatgttgcttataaaaggctgtgtatatttt
gctggtatatgaatgcacatgcattttttttttttgagacagagttgcccaggctggatt
tcagctggcatgatttcaactctctgcaacctccaccccccgggttcaagcaattctcat
gcctcagcctcccaagtagctgggactacaggcatgcgccaccacacctgactaattttt
ttttcctttttagtagagatggggtttcaccacgttggctaggctggttttgagctcctg
actgcaggttatctgcctgcctcagcctcctaaagtgctgaaattacaggcgtgggccac
tgcacatttctttttctggtctggaagcaaacccaacacaatgttaatagcagttacctt
tgaggtaaaggtttattttatttaaaattatttttactttccttatgtttgagtgttttt
taccatgtgtatattatgttacttacatttttaaagaaaaattgtcaacaaggttacctg
gatagtgaaattatgaatgatctttgctttcttatatgtttctgtttcaaaaacacagta
aatgttatttattttgtggaagatttctatcaataaaggaaaaccagagaatgtccaggc
taatggtaggagatttagtttttgggttttctggctggctggtgctggttggggtgagtc
taggctggctgctggtggtggggcagacccctgaggccctcagagtcatacacagaggaa
gagcttgggtcagactttggccttttctgcttgtttcatcaaaacagaaaccacaagctt
tatcacagcctcaaaatgcattttctctcttctttggggttaaaaacaaatttttcttca
aagaaagcatattgagatttttaagtcaagttcacaattaaaaaaaaattttcttgaaag
atgtgtgtattgtcttaacatgggttccctttgttaacacctcccagtggatttatttgg
catgagggatctaggggcaggaccctggacttcttcttagacaacctaacctctttggtc
cagaactatctcaaggttctgctaccaacatgttagaagcaaaaggcccaggaaacaacc
cagtcacttcatgggaattggcagctttaaatgtgagtacacacacagcagttggttatc
tctttgggggttaagttaccccatttagtgccgacttgtgcagacaatggggcagaggcc
agagacaccagagtaatccttttacctgcctgatctggagtgacattggaatgaaggggc
actttcagagggtatcagtttcctaggtaagttctgcaaacaagcccgcagagcctcagc
ttcctcattgtgcgtccactgcaggctgaaaaaagccagacctgcttgtcagtcccgcaa
acagatgaaagacctacttgggttcggtggaattccattgtcgtggtatttactcacagt
cgcctagcgcctcccctgaaatgtggactgcaaggggactagttttcatgccaagcacca
catcctgaaagcataaacagcgattaaaacttattataaaaagagttggaagggcagcca
gggttttccgtctctcttggtgaataaactcaactcctacatgccaacaccttgtggtta
cccaatactctctctgtcctgtcatttttctttcaaagaagccacaccttccatttctca
ctgcaaagagctgtggtgacagacatgtaagaattttattttctttttaaagtaacttag
attacagtagggctgccatgagaaaggtgacacagaagagcagacgagagggagcagaaa
ggtgtatgcagattctcttgtgtccatcagccttaagaacaggggctgctgagccaagtc
agaggctgaagccactcctagggataagatccccccatgtctctactctggggacattag
ctccactgtttagactagcttctcactttcacccacatttttaggccagtggaaacttcc
ataaatccaggtcatacattcacattcaaccgatgtgtgtttacccactacttccgatct
gaaattttgggaagctggagaacggacagggacacttgcccacagctctgcaggccagag
gtggtcccattctgggactttctgtccttctctccagctgaacaaacctgatgatgctgg
cctcctctcatagaccttttaaatgctgcatgaagtccaaaccacttagcagaattctgg
tccccaccttcacttccaacctcctggatgccctcttgccctctactctttgttctggtc
atgaaatagaaaatataatcctgcacccggtctttacctgggcagggcccctgtcctgga
atgcctcccttttcctgctttatctaactcgcctctgttcagtgaagacccaacttaaat
gccgtcttgtctgggaaatcttacttcaatcctctaggaaatattaattgcttctccctt
tagagcagattatcagtgatattatttgtttgtatgatcatctccccaaaaagcttctta
agggtttcaggaaagcattttgctcatttccatatcttcagttcctagcaatatacttgg
taatcaatatgtgtttgttgaagtttgtatgagatcatcaatatttctcatgcagagaac
agagaaaagagaacaacaataaaatatcttcaatgctgggctatcctgaaacagagattt
ttttgtttgctacatttttttttttttgagacagggtcttgctttgccacccaggctgga
gtgcagtgtcgtgatcacagctcactgcaggctctacctcctgggctcaggtgatcctcc
cacctcagcctcctggctagctgggactatagacatgtgccactatgcttagctaatttt
ttgtatttttttgttgagacaaggttttaccatgttgcccagGCTGTTCTTGAGCTGGGC
T
Primer
(+)AAGCGACTTTTGTGCCTCAG |
(+)AGCCCAGCTCAAGAACAGC |

|